Publications
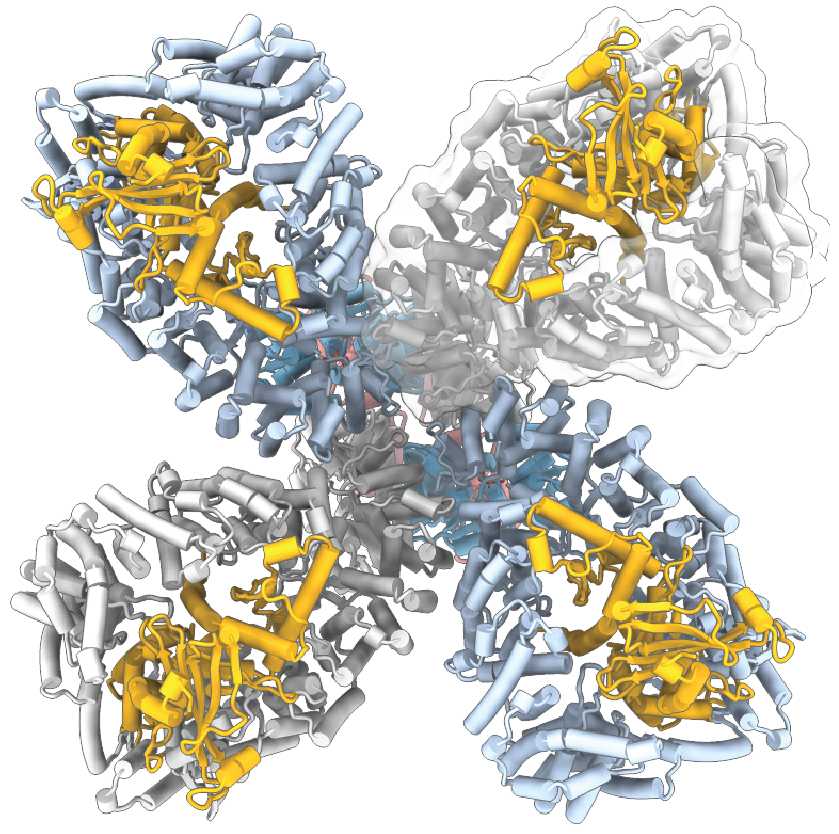
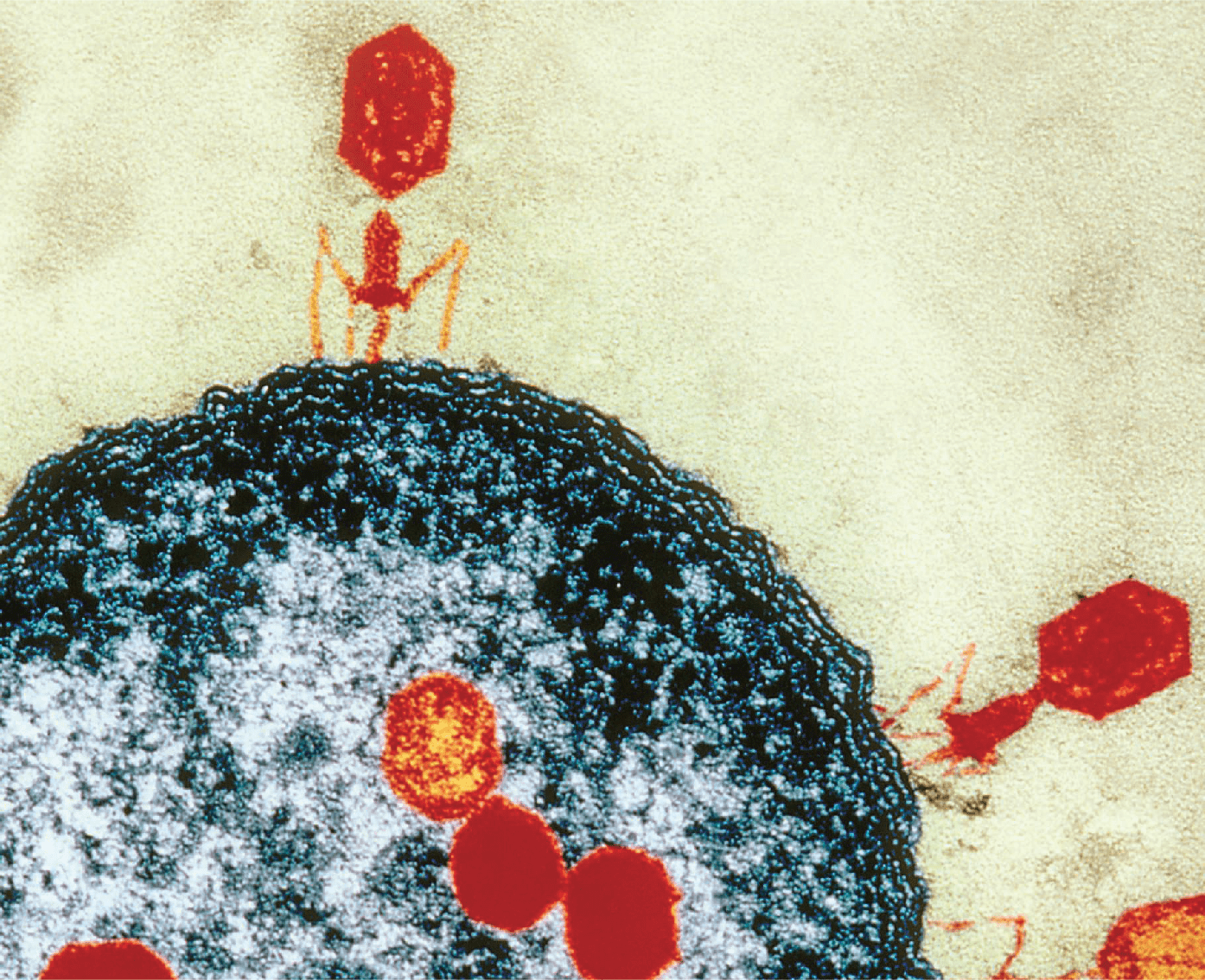
15. Gao LA*†, Wilkinson ME*, Strecker J*, Makarova KS, Macrae RK, Koonin EV, Zhang F†. Prokaryotic innate immunity through pattern recognition of conserved viral proteins. Science 377, eabm4096 (2022) (Highlighted in: Quanta Magazine, Cell Host & Microbe, Trends in Microbiology, Nature Reviews Immunology, MIT News) (PDF)
14. Mestre MR*, Gao LA*, Shah SA, López-Beltrán A, González-Delgado A, Martínez-Abarca F, Iranzo J, Redrejo M, Zhang F, Toro N. UG/Abi: A highly diverse family of prokaryotic reverse transcriptases associated with defense functions. Nucleic Acids Res, gkac467 (2022) (PDF)
13. Gao R*, Yu CC*, Gao L, Piatkevich KD, Neve RL, Upadhyayula S, Boyden ES. A highly homogeneous polymer composed of tetrahedron-like monomers for high-isotropy expansion microscopy. Nat Nanotechnol 16, 698-707 (2021) (Highlighted in: GEN, MIT News) (PDF)
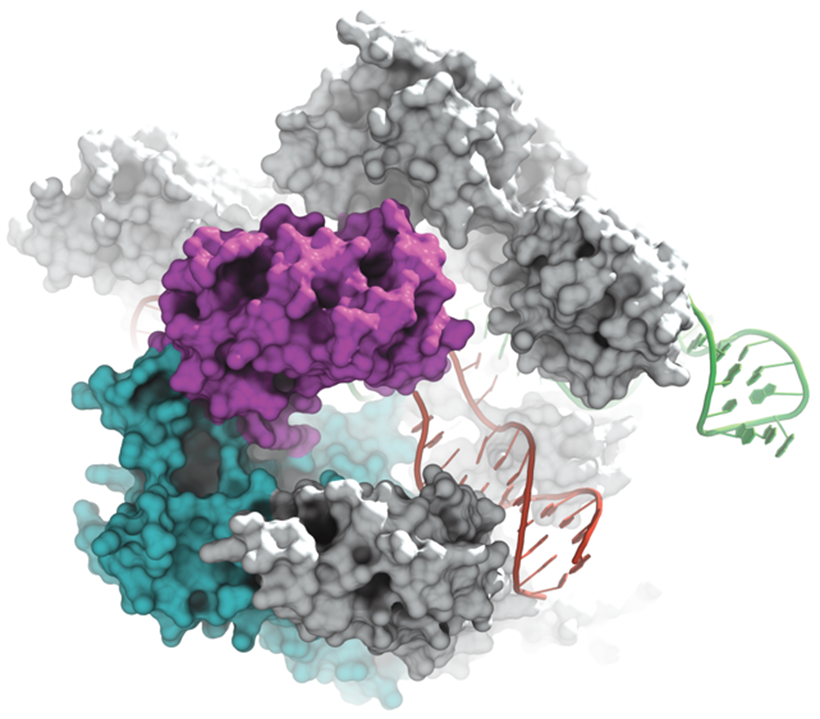
12. Gao L, Altae-Tran H, Böhning F, Makarova KS, Segel M, Schmid-Burgk JL, Koob J, Wolf YI, Koonin EV, Zhang F. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369, 1077-1084 (2020) (PDF)
11. Altae-Tran H*, Gao L*, Strecker J, Macrae RK, Zhang F. Computational identification of repeat-containing proteins and systems. QRB Discovery 1: e10, 1-12 (2020) (PDF)
10. Schmid-Burgk JL*, Gao L*, Li D*, Gardner Z, Strecker J, Lash B, Zhang F. Highly parallel profiling of Cas9 variant specificity. Mol Cell 78, 794-800 (2020) (PDF)
9. Makarova KS, Gao L, Zhang F, Koonin EV. Unexpected connections between type VI-B CRISPR-Cas systems, bacterial natural competence, ubiquitin signaling network and DNA modification through a distinct family of membrane proteins. FEMS Microbiol Lett 366, fnz088 (2019) (PDF)
8. Strecker J, Jones S, Koopal B, Schmid-Burgk J, Zetsche B, Gao L, Makarova KS, Koonin EV, Zhang F. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun 10, 212 (2019) (PDF)
7. Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OA, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259-1262 (2018) (PDF)
6. Tekin H, Simmons S, Cummings B, Gao L, Adiconis X, Hession CC, Ghoshal A, Dionne D, Choudhury SR, Yesilyurt V, Sanjana NE, Shi X, Lu C, Heidenreich M, Pan JQ, Levin JZ, Zhang F. Effects of 3D culturing conditions on the transcriptomic profile of stem-cell-derived neurons. Nat Biomed Eng 2, 540-554 (2018) (PDF)
5. Nishimasu H*, Yamano T*, Gao L, Zhang F, Ishitani R, Nureki O. Structural basis for the altered PAM recognition by engineered CRISPR-Cpf1. Mol Cell 67, 139-147 (2017) (PDF)
4. Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang F. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol 35, 789-792 (2017) (PDF)
3. Yan WX*, Mirzazadeh R*, Garnerone S, Scott DA, Schneider MW, Kallas T, Custodio J, Wernersson E, Li Y, Gao L, Fedorova Y, Zetsche B, Zhang F, Bienko M, Crosetto N. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun 8, 15058 (2017) (PDF)
2. Tillberg PW*, Chen F*, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk H, Yoshida F, DeGennaro E, Roossien DH, Desimone R, Cai D, Boyden ES. Expansion microscopy of biological specimens with protein retention. Nat Biotechnol 34, 987-992 (2016) (PDF)
1. Slaymaker IM*, Gao L*, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84-88 (2016) (Highlighted in: BBC Health, Phys.org, Nature, Nature Biotechnology, MIT News) (PDF)
(*co-first author; †corresponding author)
Bacterial pattern-recognition receptor in complex with phage portal protein